Python Basics Recap
Overview
Teaching: 0 min
Exercises: 0 minQuestions
Python Refresher
Objectives
Understanding key concepts in Python
This course follows on from the python introduction course. To ensure that we are starting from similar positions, there follows a short multiple choice quiz on key python concepts that we will be building on through this course.
Variables and Lists
1. Assigning a value to a variable
We wish to store the string Cat as a value in the variable animal, which of these lines of code will do this for us?
animal = 'Cat'animal = CatCat = animalanimal(Cat)
Solution
Answer 1 is correct
2. Assigning values to a list
We wish to create a list of values, which of these lines of code is valid to do this?
varlist = [34, 57, '2d']varlist = (12, 'vr', 95)varlist = 'xcf', 12, 97
Solution
Answer 1 is correct
3a. Indexing characters in a string
Lists and strings both contain multiple indexed values (in the case of strings these are specifically individual characters rather than other values). If we have a variable animal which contains the string penguin, which of these options will print the first character (p) for us?
print(animal[0])print(animal[1])print(animal['p'])
Solution
Answer 1 is correct
3b. Indexing characters in a string (slicing)
We can also select whole sections of lists and strings, not just single elements. Using the same variable animal, containing the string penguin, as above, which of these options will print the last 2 characters for us? (Note that there is more an one correct answer)
print(animal[5:])print(animal[6:])print(animal[6:7])print(animal[5:7])print(animal[-2:])print(animal[:-2])
Solution
Answers 1, 4, and 5 are correct
Loops
4. Constructing a for loop
Please write a simple for loop which will print out each of the characters in the animal variable one at a time.
Solution
for char in animal: print(char)
Software Modules
5. Loading new functions
We want to use the functions in the numpy library in our code. How do we open this library in our code?
import numpyload numpyopen numpy
Solution
Answer 1 is correct
If statements and conditionals
6. Conditionals
Which of these conditional tests returns a True result?
4 > 3'a' != 'b'6 >= 6.0'3' == 33 > 'c'
Solution
Answers 1, 2, and 3 return
Trueresults. 4 returnsFalse. 5 raises an Error.
7. If statements
What is printed when this if statement is used?
if 4 > 5:
print('A')
elif 4 <= 5:
print('B')
elif 4 < 5:
print('C')
else:
print('D')
ABCBandCDAandD
Solution
B. Both4 <= 5and4 < 5would return aTrueresult, but theifstatement is exited as soon as theTrueresult is returned.
Key Points
variables
lists
indexing
loops
conditionals
Dictionaries
Overview
Teaching: 0 min
Exercises: 0 minQuestions
How can I work with relational datasets?
How can I access remote data directly in my scripts?
Objectives
Learn about JSON format
How to create dictionaries
Loading JSON data and working with it
Using web API’s for accessing remote data
In previous lessons we have learnt about lists and numpy multi-dimensional arrays, which are designed for working with structured, tabular, datasets. But much of the data that we use in our day to day lives, such as that in data catalogs or the communications between modern digital services, does not fit nicely into these tabular datasets. Instead unstructured data formats, which use labels or ‘keys’ to identify each data object are needed. One of the most common formats for such data is the JavaScript Object Notation (JSON) file format. This format was originally developed to fulfil the need for a self-contained, flexible format for real-time server-to-browser communication, and is now used as the basis for many unstructured data formats.
One example of such usage in research is the storage of metadata for data, programs, workflows, or any other such object in a Research Object Crate (RO-Crate). These metadata records take the form:
{
"@context": "https://w3id.org/ro/crate/1.1/context",
"@graph": [
{
"@id": "ro-crate-metadata.json",
"@type": "CreativeWork",
"about": {
"@id": "./"
},
"conformsTo": {
"@id": "https://w3id.org/ro/crate/1.1"
}
},
{
"@id": "./",
"@type": "Dataset",
"mainEntity": {
"@id": "tracking_workflow.ga"
},
"hasPart": [
{
"@id": "tracking_workflow.ga"
},
{
"@id": "object_tracking_pipeline.png"
}
],
"author": [
],
"provider": [
{
"@id": "#project-1"
}
],
"license": "Apache-2.0",
"sdPublisher": {
"@id": "#person-1"
},
"sdDatePublished": "2021-01-01 00:00:00 +0000"
},
{
"@id": "#galaxy",
"@type": "ComputerLanguage",
"name": "Galaxy",
"identifier": {
"@id": "https://galaxyproject.org/"
},
"url": {
"@id": "https://galaxyproject.org/"
}
},
{
"@id": "#project-1",
"@type": "Organization",
"name": "Science Workflows",
},
{
"@id": "#person-1",
"@type": "Person",
"name": "Alice Smith",
}
]
}
The layout of this object is organised using key:value pairs, where the key is a unique string, and the value can be any data type, including other data structures. This simple layout allows quite complex data objects to be constructed.
Dictionary
In python this structure is implemented using the ‘dictionary’ object. Below we will go through the principles of creating and working with these objects. Then we will introduce a library for working with JSON files.
Creation
Lists are created by using square brackets [ ].
Dictionaries are created by using curly brackets { }, e.g.:
d = {}
The simplest way to create a dictionary with some value is:
d = {'keyname': 'keyvalue'}
Following the previous example, we can create a python dictionary using the name of a person as the key and their age as the value:
d = {'alice': 35, 'bob': 18}
print(d)
{'alice': 35, 'bob': 18}
Alternatively, a dictionary object can be created using the dict function, in a similar manner to using the list function. When using the dict function we need to indicate which key is associated with which value. This can be done in a number of ways, firstly with tuples:
d2 = dict([('alice', 35), ('jane', 24), ('bob',18)])
or with direct association:
d3 = dict(bob=18, alice=35, jane=24)
or using the special zip function, which can be used to create a set of tuples from the given iterable lists:
d4 = dict(zip(['jane','alice','bob'],[24,35,18]))
Accessing elements
To access an element of the dictionary we must use the key:
print('The age of alice is :', d['alice'])
The age of alice is: 35
We can also use a variable to index the dictionary:
key = 'alice'
print('The name of the person is used as key:', key)
print('The value associated to that key is:', d[key])
The name of the person is used as key: alice
The value associated to that key is: 35
Adding an element
Adding an element to a dictionary is done by creating a new key and attaching a value to it.
print('Original dictionary:', d)
d['jane'] = 24
print('New dictionary:', d)
Original dictionary: {'alice': 35, 'bob': 18}
New dictionary: {'alice': 35, 'bob': 18, 'jane': 24}
To add one or more new elements we can also use the update method:
d_extra = {'tom': 54, 'david': 87}
d.update(d_extra)
print('Updated dictionary:', d)
Updated dictionary: {'alice': 35, 'bob': 18, 'jane': 24, 'tom': 54, 'david': 87}
Dictionary Concatenate Warning
Unlike lists it is not possible to use the
+operator to concatenate dictionaries:{'alice': 35} + {'bob': 18}--------------------------------------------------------------------------- TypeError Traceback (most recent call last) <ipython-input-39-a6305e6df312> in <module> ----> 1 {'alice': 35} + {'bob': 18} TypeError: unsupported operand type(s) for +: 'dict' and 'dict'
Key Uniqueness Warning
Keys have to be unique; you cannot have two keys with the same name. If you try to add an item using a key already present in the dictionary you will overwrite the previous value.
print('Original dictionary:', d) d['alice'] = 12 print('New dictionary:', d)Original dictionary: {'alice': 35, 'bob': 18, 'jane': 24} New dictionary: {'alice': 12, 'bob': 18, 'jane': 24}
Equality between dictionaries
To be equal, all the elements which compose the first dictionary must be present in the second, and only those elements.
The position (ordering) is not important.
d1 = {'alice': 12, 'bob': 18, 'jane': 24, 'tom': 54, 'david': 87}
d2 = {'tom': 54, 'david': 87}
d3 = {'bob': 18, 'alice': 35, 'jane': 24}
d4 = {'alice': 35, 'bob': 18, 'jane': 24}
print('Dictionary 1 and dictionary 2 are equal:', d1 == d2)
print('Dictionary 1 and dictionary 3 are equal:', d1 == d3)
print('Dictionary 3 and dictionary 4 are equal:', d3 == d4)
Dictionary 1 and dictionary 2 are equal: False
Dictionary 1 and dictionary 3 are equal: False
Dictionary 3 and dictionary 4 are equal: True
Splitting out keys and values
Dictionaries have some special methods. Two of the most useful are keys and values. These return the keys and the values of the dictionary respectively.
d.keys()
dict_keys(['alice', 'bob', 'jane', 'tom', 'david'])
d.values()
dict_values([12, 18, 24, 54, 87])
Note that the dict_keys and dict_values objects are iterable but are not lists. This means that they can be used somewhere like a for loop but you can not index them directly.
d.values()[0]
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
TypeError: 'dict_keys' object is not subscriptable
If you want to index keys or values directly, you can convert them to lists with the list function.
list(d.values())[0]
12
Presence (or not) of an element inside a dictionary
It is possible to test if a key is present in the dictionary (or not) using the keyword in, just as we did at the start of this lesson for values within a list:
'alice' in d
True
'mark' in d
False
Note, however, that we can’t directly test for the presence of values:
12 in d
False
Instead we would have to use the values method to search these:
12 in d.values()
True
JSON files
Because JSON files are such a widely used format, python has a built in package for working with JSON, called json:
import json
import glob
filenames = sorted(glob.glob('*.json'))
j_objects = {}
for filename in filenames:
with open(filename) as f:
j_objects[filename] = json.loads(f.read())
The f.read method reads the file as a single string, which the json.loads() method then turns into a dictionary, following the JSON standard.
HTTP requests
Although lot of information is available on the internet for general use, without automated tools for accessing this data it is difficult to make full use of it. Python has a number of libraries for making HTTP requests, to help with this automation, of which the requests library is the most commonly used. This library provides a streamlined application process interface (API) for carrying out these tasks, and has built in JSON support, for easy digesting of the retrieved data.
The basic interaction for making a HTTP request is:
import requests
source_url='https://api.datacite.org/dois/10.48546/workflowhub.workflow.56.1'
requests.get(source_url)
<Response [200]>
The HTTP request returns a response code - a value of 200 indicates the request was successful. There are a wide range of possible response codes. Those starting as 2XX generally indicate success, whereas those starting with 4XX indicate a failure of some sort (including the most common: 404 Not Found).
The HTTP request we made returned more than just the response code, there will also be the attached content that we requested. In this case our request was made to an API which returns citation information associated with the DOI 10.48546/workflowhub.workflow.56.1.
Rather than being presented as a complex webpage, this information is returned as a machine-readable string, similar to the JSON file we read earlier, so we can read this in a similar manner:
response = requests.get(source_url)
record = response.json()
Once the data is in a dictionary we can start exploring it - first step is to check the keys available:
record.keys()
dict_keys(['data'])
The upper level of the dictionary is simply data - so we can move to the second level:
record['data'].keys()
dict_keys(['id', 'type', 'attributes', 'relationships'])
The id contains the DOI that we used to find this entry, while the attributes contains the metadata for the object referred to by the DOI. By digging further into the dictionary we can extract information about the object.
Find the title
What is the path to find the title of the object?
solution
record['data']['attributes']['titles'][0]['title']
Find the title (part 2)
Assuming that all JSON objects returned by this API follow the same layout as this record, write a simple function that will return the title of any DOI it is given.
Test that your function works using the DOI:
10.5281/zenodo.4416028solution
def doi_title( doi_string ): source_url = f'https://api.datacite.org/dois/{doi_string}' response = requests.get(source_url) record = response.json() return(record['data']['attributes']['titles'][0]['title'])Testing the function:
doi_title('10.5281/zenodo.4416028')'Britain Breathing 2016-2019 Air Quality and Meteorological Dataset'
Speed Tests…
Sequences are a great tool but they have one big limitation. The execution time to find one specific value inside is linear, as can be shown by fruitless searches for the string x within increasingly long lists of integers.
short_list = list(range(100_000))
long_list = list(range(1_000_000))
We use the built-in %timeit function, to test the speed of these searches:
%timeit -n100 'x' in short_list
2.02 ms ± 529 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
%timeit -n100 'x' in long_list
17.4 ms ± 1.35 ms per loop (mean ± std. dev. of 7 runs, 100 loops each)
Note that the increase in search time is (very roughly) linear.
This is a real problem because the membership test is a very useful and common procedure. So we would like to have something which is not dependent on the number of elements.
Testing access time for large dictionaries
Create two dictionaries, one with 100,000 key:value pairs, the other with 1,000,000 key:value pairs, using the lists created at the start of this lesson. Then use these to test the access times for dictionaries using the
%timeitfunction. How do the access times compare with those for the lists, are they quicker or slower, and do the access times scale linearly with the size of the dictionary?Solution
The dictionaries can be created using the
zipmethod:short_hash = dict(zip(short_list,short_list)) long_hash = dict(zip(long_list,long_list))The access times for the dictionaries are 100-1,000 times faster than for lists, and the search time does not increase with dictionary size.
%timeit -n100 'x' in short_hash34.2 ns ± 2.31 ns per loop (mean ± std. dev. of 7 runs, 100 loops each)%timeit -n100 'x' in long_hash48.4 ns ± 5.39 ns per loop (mean ± std. dev. of 7 runs, 100 loops each)Note: it is likely that your first report back from the test will look like this:
The slowest run took 308.65 times longer than the fastest. This could mean that an intermediate result is being cached. 2.52 µs ± 5.91 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)This is because the secret behind the speed of searching a dictionary is the caching of the keys after the first access to them. Running the tests a second time will give above test results.
This caching behaviour is very useful for datasets for are accessed regularly.
Key Points
JSON is simple
Dictionaries are defined using
key:valuepairsDictionaries can be nested, and mixed with lists
Web API’s can be accessed using the
requestslibrary
Numpy and Matplotlib Essential
Overview
Teaching: 0 min
Exercises: 0 minQuestions
How do I select data within tables for processing?
How can I load different data formats?
Objectives
Learn about NumPy matrix objects
Learn about masking datasets
Learn about specialised data formats, and how to use them
NumPy extends the Python language by providing new types (array, matrix, masked_array…), functions and methods to realise efficient numerical calculation using Python. NumPy is the numerical library for Python. It is too big to be covered in one day, so today we will learn a few of the basic objects and functions.
NumPy is a foundation block for many other python libraries used in science and data science, e.g.:
- scipy Fundamental library for scientific computing (interation, optimisation…)
- pandas data structure and data analysis tools
- matplotlib Python 2D plotting
And more specialised libraries such as:
- astropy for astronomy
- h5py to interact with HDF5 format datafiles
- scikit-learn for Machine Learning
- TensorFlow for Deep Learning
We are going to learn some of the basic commands not seen in the first course. We will introduce some of these libraries but keep in mind that we are just covering the basics so you can understand how to start using this library. If you find that your interest is piqued and/or they are applicable to your problem, you can use the documentation to learn more.
The array is the central object of NumPy. It is a multi-dimensional object which contains numerical data. The main difference between NumPy arrays and Python lists is the set of operations that can be performed on them. In this lesson we are going to learn a little more about how to use this fundamental library to do any numerical analysis in Python.
NumPy arrays are good for arrays of numerical data where each dimension is of a consistent size. Where you want to work with heterogeneous data and performance is not a problem, working with Python lists and dictionaries is easier than working with uneven datasets in NumPy. Similarly, while NumPy arrays do technically support having different data types in different columns, it is awkward to do and breaks a lot of the useful NumPy methods. It is best to use libraries for the specific purpose for which they are designed, so any sort of tabular data is better handled with something like Pandas.
To start, we are going to import the two libraries numpy and matplotlib that will be used in this episode.
import numpy as np
import matplotlib.pyplot as plt
Here we import the libraries using a common shorthand for them, np for numpy, and plt for matplotlib.pyplot. You will encounter this in many python scripts using these libraries, and we will continue to use these shorthands below.
If we wanted to create a list of odd numbers in base Python we would use range and list:
odds = list(range(1, 10, 2))
print(f'odds is type {type(odds)}, and contains {odds}')
odds is type <class 'list'>, and contains [1, 3, 5, 7, 9]
To create the same list as a NumPy array we can use the np.arange function:
odds_np = np.arange(1, 10, 2)
print(f'odds_np is type {type(odds_np)}, and contains {odds_np}')
odds_np is type <class 'numpy.ndarray'>, and contains [1 3 5 7 9]
Note that we did not need to convert the output of np.arange, as we did for the output of range, because np.arange returns a NumPy array directly, rather than returning a range object.
NumPy Array Addition
What is the difference between addition of NumPy arrays and lists? What is the result of these two additions?
odds + odds odds_np + odds_np
[1, 3, 5, 7, 9, 1, 3, 5, 7, 9]for both[1, 3, 5, 7, 9, 1, 3, 5, 7, 9]for odds, and[2, 6, 10, 14, 18]for odds_np[2, 6, 10, 14, 18]for odds, and[1, 3, 5, 7, 9, 1, 3, 5, 7, 9]for odds_np[2, 6, 10, 14, 18]for bothSolution
The correct answer is (2)
Reshaping NumPy Arrays
NumPy arrays are multidimensional objects for storing data (not necessarily numerical). The shape of the array can be modified using the method reshape:
arr = np.arange(1, 18, 2)
arr_2d = arr.reshape((3, 3))
print(f'arr has shape {arr.shape}, and contains:')
print(arr)
print(f'arr_2d has shape {arr_2d.shape}, and contains:')
print(arr_2d)
arr has shape (9,), and contains:
[ 1 3 5 7 9 11 13 15 17]
arr_2d has shape (3, 3), and contains:
[[ 1 3 5]
[ 7 9 11]
[13 15 17]]
Checking and changing the type of the data within the array is relatively straightforward:
print(arr.dtype)
arr = arr.astype(float)
print(arr.dtype)
int64
float64
Working with Matrices
NumPy arrays are not matrix objects, but NumPy does provide a matrix object, which has the characteristics of a mathematical matrix:
mat = np.matrix(arr_2d)
type(mat)
numpy.matrix
These can be used to carry out matrix multiplication:
mat * mat
matrix([[ 87., 105., 123.],
[213., 267., 321.],
[339., 429., 519.]])
Masked Arrays
A very useful tool that NumPy provides for working with experimental data is the masked array. When you are taking data from an experiment you always have some data which are not present, or with a bad signal to noise ratio, or that you cannot use for some other reason. This function allows you to select which data you wish to work with within your dataset.
Masked arrays associate a NumPy array with another array composed only of boolean values (True or False). These tell NumPy whether to use (or not) the respective element.
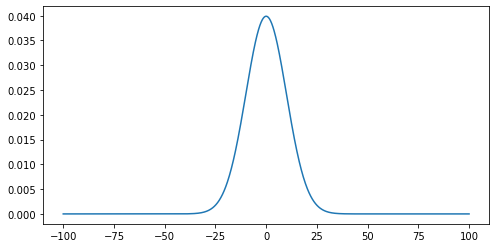
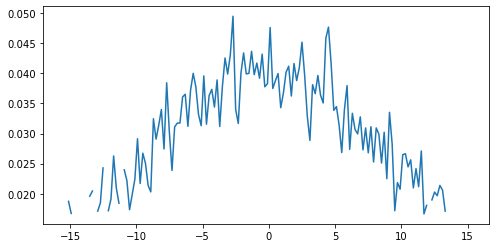
To demonstrate this we are going to create a Gaussian function and use it to generate an example dataset and generate a plot. We will then add some noise to it and use a masked array to filter out the noisy data. This represents the kind of processing that can be used for datasets such as a seismographs, where we would wish to isolate single events from noisy background data.
Reminder: the Gaussian function is defined by:
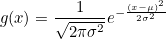
Gaussian Function
- Create a function called
gausswhich will take three arguments (inputs): x, µ, and σ, as defined above. (x is an array, µ is the position of the centre of the curve/peak and σ is the width of the bell)- Create a NumPy array using the ‘numpy’ function ‘linspace’ which will contain 1000 points equally spaced between x=-100 and x=100. Hint: You can print the help documentation of a function with ‘help(name_of_the_function)’
- Using the above gauss function and the array, create a list which contains the value of the gauss from x=-100 to x=100.
- Use the ‘matplotlib’ library to plot the curve with mu=0 and sigma=10.
challenge 1:
def gauss(x, mu=0, sigma=1): return (1. / (np.sqrt(2 * np.pi * sigma ** 2)) * np.exp( - (x - mu) ** 2 / (2 * sigma ** 2)))challenge 2:
x = np.linspace(-100, 100, 1000)challenge 3:
mu = 0 # the position of the center of the peak sigma = 10 # The width of the 'bell' g = gauss(x, mu, sigma)challenge 4:
plt.plot(x, g) plt.show()
Noisy Signal
Now we are going to add some random noise to that curve. To do it we can use the NumPy function normal from the module random provided by NumPy library. We will scale the magnitude of the noise so it is (roughly) a 10th of the magnitude of the Gaussian maximum:
noisy = np.random.normal(g, scale=g.max() / 10)
plt.plot(x, noisy)
plt.show()
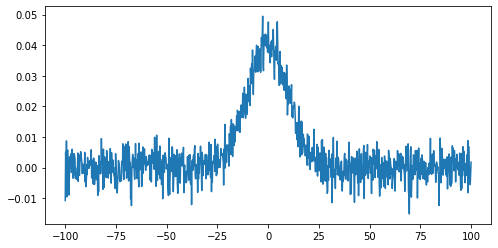
To identify any signal in the data we can use the standard deviation as an estimate of the noise around the mean value of the data.
stddev_noisy = np.std(noisy)
mean_noisy = np.mean(noisy)
print(f'standard deviation is: {stddev_noisy}')
print(f'mean value is: {mean_noisy}')
standard deviation is: 0.011592652442611553
mean value is: 0.005047252119578472
We will create a mask for the data, by selecting all data points below this threshold value (we’ll assume here that any signal we might be interested in is positive):
mask = noisy < (stddev_noisy + mean_noisy)
This creates an array of boolean values, the same shape as our original data, with True values where the conditional statement has been met.
The mask and noisy data can now be combined using the array function of the NumPy masked arrays module (np.ma). Any True value in the mask will exclude the corresponding element from subsequent computation or plotting:
noisy_ma = np.ma.array(noisy, mask=mask)
plt.plot(noisy_ma)
plt.show()
Working with images
We are going to learn some commands that deal with images. Since most scientific domains use their own file format, we obviously can not learn all of them. We will use a typical astronomical image format: the ‘fits’ file.
In the data directory you should find a file called 502nmos.fits.
We need to use the astropy I/O library fits to open this file. This can be imported as:
import astropy.io.fits as pyfits
The file can now be opened using:
im1 = pyfits.open('data/502nmos.fits')
Memory management for large files
By default pyfits opens a file with the option
memmap=True. This option opens the fits file without copying the data into memory and allows us to open very large files which will not fit into physical memory.
Fits files are composed of a list of HDUs (Header and data units). We can list the information with the method info.
im1.info()
Filename: data/502nmos.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 290 (1600, 1600) float32
1 502nmos_cvt.tab 1 TableHDU 353 1R x 49C [D25.17, D25.17, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, A1, E15.7, I12, I12, D25.17, D25.17, A8, A8, I12, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, I12, I12, I12, I12, I12, I12, I12, I12, A48, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7, E15.7]
Here we’re only interested in the primary HDU which is an image and is called PRIMARY.
The base file system can be accessed in the same manner as a dictionary - using either the key im1['PRIMARY'] or the index im1[0].
The image comprises of two parts: a header, and then the image data. The header can be accessed using the header attribute:
im1['PRIMARY'].header
SIMPLE = T / FITS STANDARD
BITPIX = -32 / FITS BITS/PIXEL
NAXIS = 2 / NUMBER OF AXES
NAXIS1 = 1600 /
NAXIS2 = 1600 /
EXTEND = T / There maybe standard extensions
BSCALE = 1.0E0 / REAL = TAPE*BSCALE + BZERO
BZERO = 0.0E0 /
OPSIZE = 2112 / PSIZE of original image
ORIGIN = 'STScI-STSDAS' / Fitsio version 21-Feb-1996
FITSDATE= '2005-07-01' / Date FITS file was created
FILENAME= '502nmos_cvt.hhh' / Original filename
...
To access the data we use the data attribute:
imdata = im1['PRIMARY'].data
type(imdata)
<class 'numpy.ndarray'>
Note that this data is in the form of a NumPy array, and so we can use our standard NumPy tools for processing and displaying this data.
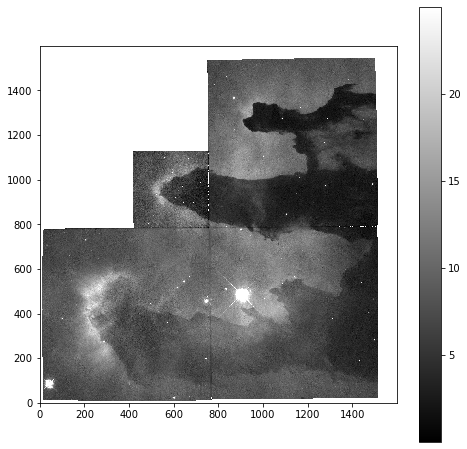
To start with we will look at the unprocessed image of the nebulae:
plt.imshow(imdata, origin='lower', cmap='gray')
plt.colorbar()
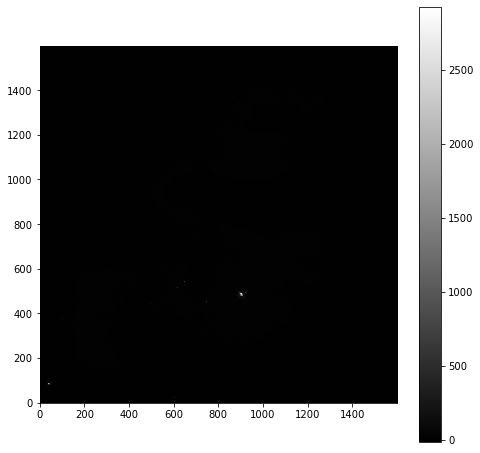
As is common for astronomical images, it is difficult to see anything on this image because a few very bright objects within the frame have saturated the CCD, and so a linear output shows only a limited number of pixels.
To improve the visible output we will carry out some simple analysis of the image, so that we can solve this contrast problem.
First we examine the general stats of the data (using built-in methods, except for the median, which has to be called from NumPy directly):
print('mean value im1:', imdata.mean())
print('median value im1:', np.median(imdata))
print('max value im1:', imdata.max())
print('min value im1:', imdata.min())
mean value im1: 6.237272
median value im1: 6.7385635
max value im1: 2925.8718
min value im1: -12.439324
These show that, while the range is very large, the mean and median values are both very low, suggesting that there are only a few pixels with very high values.
This can be confirmed by plotting a histogram of the number of pixels vs the number of photons:
hist = plt.hist(imdata.ravel(), bins=100)
plt.xlabel('Number of Photons')
plt.ylabel('Pixel Count')
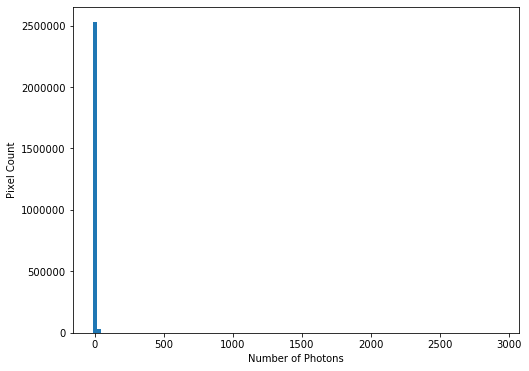 This confirms our suspicions that many pixels have very low photon counts.
This confirms our suspicions that many pixels have very low photon counts.
Flattening N-D arrays
To plot a histogram of all the data we needed to flatten the two dimensional data array. This was done using the built-in
ravelfunction.
To see the full range of photon counts we will use a log scale:
hist = plt.hist(imdata.ravel(), bins=100)
plt.yscale('log')
plt.xlabel('Number of Photons')
plt.ylabel('Pixel Count')
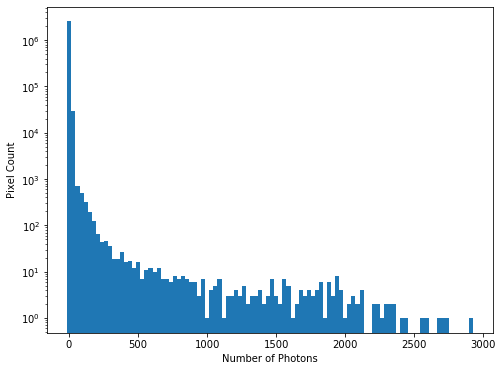
While almost all pixels have a photon count of zero (or near zero), only a few (<1000) photon counts are above 100. There does appear to be a significant number of pixels with values between these two extremes though, so we will constrain the range of our x-axis to between the limits of 1 and 30, to better see these data. The limit of 30 is obtained through previous analysis - if you were carrying out this investigation of the data yourself you would use trial and error to find the best cut-off point for this plot.
hist = plt.hist(imdata.ravel(), bins=100, range=(1,30))
plt.xlabel('Number of Photons')
plt.ylabel('Pixel Count')
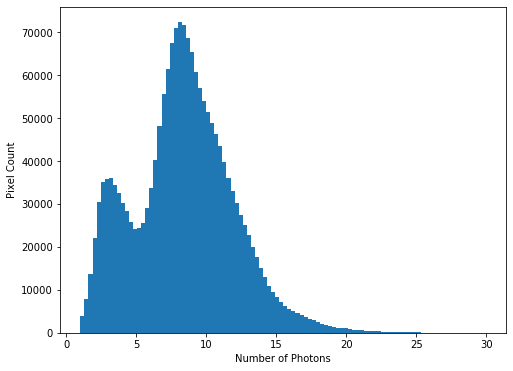 We see that there is a bi-modal distribution, with the largest peak around 8-9 photons, and a smaller peak around 3-4 photons.
We see that there is a bi-modal distribution, with the largest peak around 8-9 photons, and a smaller peak around 3-4 photons.
Now that we can see that the majority of the data is below a photon count of 25, we can start manipulating our image plot to see more details of the nebulae:
plt.imshow(imdata, origin='lower', cmap='gray', vmax=25)
plt.colorbar()
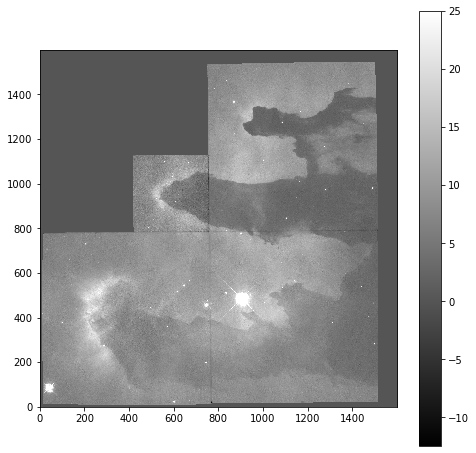
Masked Arrays (part 2)
Rather than applying the filtering of our data within the plotting tool, we can instead use masked arrays to filter our data. This allows for more nuanced filtering of the data.
We begin with creating a mask to remove values greater than 25:
immasked = np.ma.masked_greater(imdata, 25)
The immasked array contains the imdata data (you can test this by plotting immasked using imshow as we did at the start of this section). It also contains an attribute mask, which is applied to filter the data when it is plotted. We can plot this directly to see what is being removed:
plt.imshow(immasked.mask, cmap='gray', origin='lower')
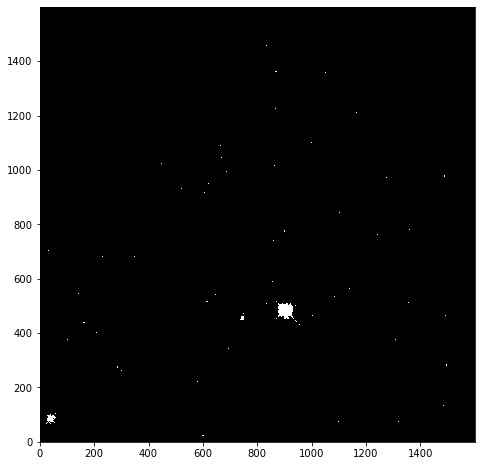
This mask is applied to the data for all built-in functions. But where we have to directly use a NumPy function we have to make sure we use the equivalent function in the mask (ma) library:
print('original average:', imdata.mean())
print('Masked average:', immasked.mean())
print()
print('original max:', imdata.max())
print('Masked max:', immasked.max())
print()
print('original min:', imdata.min())
print('Masked min:', immasked.min())
print()
print('original median:', np.mean(imdata))
print('Masked median:', np.ma.median(immasked))
original average: 6.237272
Masked average: 5.976717048232941
original max: 2925.8718
Masked max: 24.981937
original min: -12.439324
Masked min: -12.439324
original median: 6.237272
Masked median: 6.725283622741699
Removing negative photon counts
Photon counts cannot be negative, but some of our data is (some is due to no measurements being taken, and some is also likely due to sampling errors). So we would like to remove these erroneous data from our plot too.
- Create a new masked array (
immasked2) where the lower limit is set to zero (you can use thenp.ma.masked_less_equalfunction to do this).- Update the immasked2.mask to apply both masks (you can use the logical OR operator
|to do this). Then plot the new masked data array.New mask
immasked2 = np.ma.masked_less_equal(imdata, 0)Combining masks
immasked2.mask = immasked2.mask | immasked.maskplt.imshow(immasked2, cmap='gray', origin='lower') plt.colorbar()
Key Points
NumPy arrays are not matrix objects
Array masks can be created using conditional statements
NumPy arrays can be masked to hide data you don’t want to include in an analysis
NumPy libraries are available for reading a lot of different file formats
Software Package Management
Overview
Teaching: 0 min
Exercises: 0 minQuestions
How can I load new python libraries?
How can I deal with libraries which have competing software requirements
How can I record the python libraries that I use for my work?
Objectives
Learn how to install a conda package
Learn how to use different conda channels
Learn how to create a new conda environment
Learn how to activate / deactivate conda environments
Learn how to record the setup of a conda environment
Virtual environments are a useful tool for isolating and managing the software packages you use. Doing this enables you to track the packages you use for your work (enabling you to create reproducable environments, so that others can also use your code). It also allows you to use packages which might have conflicting requirements (or even different versions of the same package) without the hassle of haveing to install and uninstall these each time.
Python has a built in tools for managing virtual environments (venv) and packages (pip). However we will not cover these tools today. While venv and pip are very useful for managing pure python packages, they are not very flexible when we want to use packages which are built on other languages (or perhaps do not use python at all). Another tool, conda, has been built to extend their functionality to cover these mixed-language packages, for a wide range of computing platforms, and it is this which we will cover today. More details on the differences between conda and pip are given in this summary.
The lesson below is not conducted using a python interpreter, but instead using the unix shell. When you are asked to type in code below, please do this in a terminal window and not in this browser.
Installing Conda
The conda package manager is available to install by itself (using the miniconda installer), or with a preprepared set of commonly used packages and a graphical user-interface (GUI) for managing your packages (using the anaconda installer). Miniconda is a good choice if you have limited bandwidth or disk space, and are happy to install packages as you need them using the command line interface (CLI). Anaconda is a good choice if you have more disk space (it requires >3Gb) and want to use a GUI.
We will not cover installation in this lesson but more information is available on both of these conda installers, as well as links for downloading the installers, here: https://docs.conda.io/projects/conda/en/latest/user-guide/install/download.html.
Package Channels
Conda uses “channels”, locations where software packages are stored. Conda has a default set of these channels that it searches when you request a package or update. You can modify this list of channels, to add other public or private channels that you wish to use. One of the most commonly added is conda-forge, a community channel with thousands of contributors who use its unified, automated build infrastructure to share their software with the world. Another widely-used channel is bioconda, which is dedicated to bioinformatics software.
We can see what channels are being used by the command:
conda config --show channels
This will (likely) list the the following:
channels:
- defaults
The channel order is important for determining priority when installing packages. Those from channels higher in the list will be given priority over those lower in the list. After this the newer versions of a package are given priority over older versions of a package (and then build numbers, for a given version). The order in which priority is determined can be changed, we won’t cover this today, but more information can be found in the conda managing channels documentation.
To add a channel we can use:
conda config --add channels conda-forge
conda config --show channels
channels:
- conda-forge
- defaults
We can repeat this to add as many channels as we need:
conda config --add channels bioconda
conda config --show channels
channels:
- bioconda
- conda-forge
- defaults
This adds the new channel to the start of the list. Which gives that channel highest priority. The documentation for bioconda, however, recommends a priority order of conda-forge, bioconda, and finally defaults. To correct the channel order we can simply re-add the conda-forge channel:
conda config --add channels conda-forge
Warning: 'conda-forge' already in 'channels' list, moving to the top
When we do this we should be warned the channel is already in the list, but it is being moved up to the highest priority position.
removing or appending channels
Channels can be removed using the
--removeflag instead of--add. To add a channel to the end of the list, rather than the front, we can use--appendinstead of--add(and, if we wish to be explicit in our intent to put a channel at the front of the list we can use--prepend, which behaves in the exact same way as--add).
Searching for Packages
Once you’ve added the channels you wish to use, you can now search them for the software that you need. This is simply carried out using conda search. For example, if we wished to find out what versions of Gromacs are available, we can use:
conda search gromacs
# Name Version Build Channel
gromacs 4.6.5 0 bioconda
gromacs 2018.3 h470a237_0 bioconda
gromacs 2018.4 h470a237_0 bioconda
gromacs 2018.5 h04f5b5a_0 bioconda
gromacs 2018.6 h04f5b5a_0 bioconda
gromacs 2018.6 h04f5b5a_1 bioconda
gromacs 2018.6 hd895feb_2 bioconda
gromacs 2019 h04f5b5a_0 bioconda
gromacs 2019.1 h04f5b5a_0 bioconda
gromacs 2019.1 h04f5b5a_1 bioconda
gromacs 2019.1 h04f5b5a_2 bioconda
gromacs 2020.5 hd895feb_1 bioconda
gromacs 2020.5 hd895feb_2 bioconda
gromacs 2020.5 hd895feb_3 bioconda
gromacs 2020.5 hd895feb_4 bioconda
gromacs 2020.5 hd895feb_5 bioconda
gromacs 2020.6 h94ec9d8_0 bioconda
gromacs 2021 hd895feb_0 bioconda
gromacs 2021 hd895feb_1 bioconda
gromacs 2021.1 hd895feb_0 bioconda
This shows you the name of the package you searched for, then the version of the software, build number (which can increment up as bugs are fixed, see above for version 2020.5), and the channel that the package is in.
If we know the version of the software we are interested in we can narrow this search down. For example, if we are only interested in gromacs versions greater than 2020 we can use:
conda search "gromacs>=2020"
# Name Version Build Channel
gromacs 2020.5 hd895feb_1 bioconda
gromacs 2020.5 hd895feb_2 bioconda
gromacs 2020.5 hd895feb_3 bioconda
gromacs 2020.5 hd895feb_4 bioconda
gromacs 2020.5 hd895feb_5 bioconda
gromacs 2020.6 h94ec9d8_0 bioconda
gromacs 2021 hd895feb_0 bioconda
gromacs 2021 hd895feb_1 bioconda
gromacs 2021.1 hd895feb_0 bioconda
Note that we’ve put the search string in quotations, in order to ensure it is passed to conda correctly.
We can also use wildcards in the version string. For example, if we wish to find all numpy packages with version 1.20:
conda search "numpy==1.20.*"
# Name Version Build Channel
numpy 1.20.0 py37h3795f5d_0 conda-forge
numpy 1.20.0 py37ha9839cc_0 conda-forge
numpy 1.20.0 py38h64deac9_0 conda-forge
numpy 1.20.0 py39h3c955ea_0 conda-forge
numpy 1.20.1 py37h3795f5d_0 conda-forge
numpy 1.20.1 py37h43259c0_0 pkgs/main
numpy 1.20.1 py37ha9839cc_0 conda-forge
numpy 1.20.1 py37hd6e1bb9_0 pkgs/main
numpy 1.20.1 py38h43259c0_0 pkgs/main
numpy 1.20.1 py38h64deac9_0 conda-forge
numpy 1.20.1 py38hd6e1bb9_0 pkgs/main
numpy 1.20.1 py39h3c955ea_0 conda-forge
numpy 1.20.1 py39h43259c0_0 pkgs/main
numpy 1.20.1 py39hd6e1bb9_0 pkgs/main
numpy 1.20.2 py37h84c02c4_0 conda-forge
numpy 1.20.2 py37hc415c66_0 conda-forge
numpy 1.20.2 py38had91d27_0 conda-forge
numpy 1.20.2 py39h7eed0ac_0 conda-forge
Note that the build string now starts with pyXX, where XX indicates the version of python the package was built against. When conda packages are dependent on python then they usually will be built against a number of different versions of python - which gives more flexibility when trying to create an environment. Also note that two channels are listed here, the default pkgs/main and conda-forge. These duplicate some versions, but also note that conda-forge has the most recent version of the numpy software, whereas the default channel does not. conda-forge tends to be updated more frequently than the default channels.
Creating Environments
As the range and variety of software used increases, it is becoming very common for users require a number of different software programs for their work which need access to different versions of the same software library. In addition there could be conflicts between the operating system requirements (as these tend to change slowly, and be built on reliable software libraries) and the latest version of the required software package. Usually, installing all this software on the same system would be very difficult, but if we use virtual environments to separate these programs and their dependencies, then the task becomes a lot more simple.
We will start with listing the current environments:
conda env list
# conda environments:
#
base * /Users/mbessdl2/anaconda3
This shows you the environments you have installed, and the path where they are installed. In addition the * indicates which environment you currently have loaded. In the example above, anaconda is installed by the user, and is only accessible by that particular user. However, if anaconda has been installed by the administrator of that computer then you might something like:
# conda environments:
#
base * /opt/apps/anaconda
In this case the anaconda install will be accessible for all users, however no-one other than the administrator will be able to install new packages into the base environment.
You can list the packages you have installed in your current environment using:
conda list
# packages in environment at /Users/mbessdl2/anaconda3:
#
# Name Version Build Channel
_anaconda_depends 2019.03 py37_0
_ipyw_jlab_nb_ext_conf 0.1.0 py37_0
alabaster 0.7.12 py_0 conda-forge
anaconda custom py37_1
anaconda-client 1.7.2 py_0 conda-forge
anaconda-navigator 1.9.12 py37_0
anaconda-project 0.8.3 py_0 conda-forge
...
zip 3.0 h1de35cc_1 conda-forge
zipp 3.1.0 py_0 conda-forge
zlib 1.2.11 h0b31af3_1006 conda-forge
zstd 1.4.4 he7fca8b_1 conda-forge
When you begin using conda, as you can see, you are given a default environment named base. Avoid installing programs into your base environment, it is better to create separate environments to keep your programs isolated from each other. A new conda environment can be created using the command:
conda create --name myenv
This will create an environment named myenv (once you press [y] to proceed). Check this using the conda env list command.
# conda environments:
#
base * /Users/mbessdl2/anaconda3
myenv /Users/mbessdl2/anaconda3/envs/myenv
This environment will be empty of all packages though, as we can show by specifying the myenv environment when we list the installed packages:
conda list myenv
# packages in environment at /Users/mbessdl2/anaconda3:
#
# Name Version Build Channel
We can also specify the packages we want to install when creating an environment. When this is done conda will work out all the necessary supporting packages for you. For example, we will create the myenv environment again, this time installing the spyder IDE package, as well as the pandas and matplotlib packages, which we will need later:
conda create --name myenv spyder pandas matplotlib
This will warn you that you are about to overwrite another environment, press [y] to continue. Conda will then workout the new environment setup (which can take a little time), and then will list the packages which are to be downloaded, and those which will be installed, before asking if you wish to continue. Press [y] to continue, and then check that the environment exists and that the packages you expect are installed using conda env list, and conda list myenv as before.
Activating and Deactivating Environments
Now that we have installed spyder we would like to run it. However it is not available to us yet:
which spyder
(if this cannot find the spyder program it will return nothing)
Before we can use this software, we must activate the environment it is in. This can be done using:
conda activate myenv
Once this done you should see the name of the environment you are using in brackets before your command prompt. (If you have used pip, this will be familiar.) And you will now be able to access the program:
which spyder
/Users/mbessdl2/anaconda3/envs/myenv/bin/spyder
What if conda hasn’t been configured yet?
It is possible that
conda activate <ENV>will give you this error:CommandNotFoundError: Your shell has not been properly configured to use 'conda activate'. To initialize your shell, run $ conda init <SHELL_NAME> Currently supported shells are: - bash - fish - tcsh - xonsh - zsh - powershell See 'conda init --help' for more information and options. IMPORTANT: You may need to close and restart your shell after running 'conda init'.This will often happen on computers with a central installation of anaconda, where the anaconda environment management tools will have not been initialised (as many users will not need them). This can be corrected by following the instructions in the error message (assuming that your current shell is
bash, change the last word if not):conda init bashThis will modify your personal shell configuration (in file
/home/mbessdl2/.bashrcor similar). You will have to close that shell, and then open another shell for the changes to take effect.
Installing Software into an Existing Environment
During your work you often will find that you need to add new software packages to your environment. For example, we will need the astropy and pint packages for the units and quantities lesson, and so we should add these now.
To install these packages (into the current environment we are in) we can use:
conda install astropy pint
If you are installing packages into an environment which is not active (or if you want to be certain that you are installing into the correct environment), you can add -n <ENV>:
conda install -n myenv astropy pint
Uninstalling software can be carried out in a similar manner, using conda uninstall.
Recording and Reproducing Environment Setups
A big advantage of using virtual environments is the ability to record the environment in which you carry out your work, so that this can be shared with others in order to help them replicate your work.
The most basic way to obtain this information is using:
conda list --export
# This file may be used to create an environment using:
# $ conda create --name <env> --file <this file>
# platform: linux-64
_libgcc_mutex=0.1=conda_forge
_openmp_mutex=4.5=1_gnu
alabaster=0.7.12=py_0
alsa-lib=1.2.3=h516909a_0
appdirs=1.4.4=pyh9f0ad1d_0
argh=0.26.2=pyh9f0ad1d_1002
arrow=1.1.0=pyhd8ed1ab_1
astroid=2.5.6=py39hf3d152e_0
...
zeromq=4.3.4=h9c3ff4c_0
zipp=3.4.1=pyhd8ed1ab_0
zlib=1.2.11=h516909a_1010
zstd=1.4.9=ha95c52a_0
This contains similar information to the standard conda list command, but in a machine readable formation. The output information can be saved to a text file using conda list --export > envfile.txt, which can then be used to recreate the environment elsewhere, using the command conda create --name <env> --file <this file>. However, it should be noted that this will be restricted to the operating system it is created on, and the channel information is not recorded in this output - so if you have used more than just the default and conda-forge channels it may be difficult for someone else to replicate.
A better method is to use:
conda env export
name: myenv
channels:
- conda-forge
- bioconda
- defaults
dependencies:
- _libgcc_mutex=0.1=conda_forge
- _openmp_mutex=4.5=1_gnu
- alabaster=0.7.12=py_0
- alsa-lib=1.2.3=h516909a_0
- appdirs=1.4.4=pyh9f0ad1d_0
- argh=0.26.2=pyh9f0ad1d_1002
- arrow=1.1.0=pyhd8ed1ab_1
- astroid=2.5.6=py39hf3d152e_0
...
- zeromq=4.3.4=h9c3ff4c_0
- zipp=3.4.1=pyhd8ed1ab_0
- zlib=1.2.11=h516909a_1010
- zstd=1.4.9=ha95c52a_0
prefix: /home/mbessdl2/.conda/envs/myenv
This will present the same information, but in the more readable YAML format, and it will also include the channel information in the output. This information can be saved as a text file as before, and used to create a new environment using the command conda env create. However it is still operating system specific information.
To create a higher-level record of the environment you can use:
conda env export --from-history
name: myenv
channels:
- conda-forge
- bioconda
- defaults
dependencies:
- pandas
- spyder
- astropy
- pint
prefix: /home/mbessdl2/.conda/envs/myenv
This builds a list of only the packages that you explicitly requested to be installed in that environment. Where you have specified a particular version of a package, then this will be recorded, but otherwise the version number will be unconstrained. If you wish to constrain the version of a package (for example, if a certain feature you use has been removed in more recent versions of the package), then you will need to edit this information to specify that (using the information given by the standard conda list command).
To create a new environment from either of these YAML environment files, you can use the command:
conda env create --file <env file> --name <env name>
The name that you give the environment does not have to be the same as the name specified in the file, but if you don’t explicitly specify a name then the name in the file will be used.
Key Points
conda virtual environments are useful for installing programs with differing requirements
conda config --add channels <channel>adds a new channel to your list of sources
conda search <package>will find all available versions of a package in your list of sources
conda create -n <env> <package(s)>can be used to create a virtual environment from a list of packages
conda install -n <env> <pacakge(s)>installs packages in a pre-existing environment
conda activate <env>activates the named environment, giving access to the software installed there
conda deactivatedeactivates the current environment
conda env export --from-history > <file.yml>creates a portable record of your current environment
conda env create --file <file.yml> <env>creates a new environment from an environment file
Defensive Programming
Overview
Teaching: 0 min
Exercises: 0 minQuestions
How can I catch errors in my data?
How can I make best use of error messages?
Objectives
Learn how to catch and deal with errors within your code.
Learn how to catch problematic data before it is used in your code.
In this episode we are going to learn about error handling, a python structure (also common in other programming languages) which will help us to properly manage the anomalies that can be encountered when executing our code.
In science most developers write code for themselves and do not feel the need to use error handling. However, nowadays scientists have to share their work as more funding agencies are also asking for the code used in experiments to be published.
More importantly, applications are not expected to crash and we are going to learn how to avoid this non desirable behaviour.
What is the problem with this test?
Please look at the following code. Can you find the fundamental problem in this test?
val = 1 if val>0 and val<10: print('Value: ', val, 'is a digit.') elif val==0: print('Value ', val, 'is nul') else: print('Value: ', val, 'is a number.')Solution
The test assumes that
valis a number, and throws an uncontrolled error if it is not.val = 'a' if val>0 and val<10: print('Value: ', val, 'is a digit.') elif val==0: print('Value ', val, 'is nul') else: print('Value: ', val, 'is a number.')--------------------------------------------------------------------------- TypeError Traceback (most recent call last) <ipython-input-2-99c0e25bf5e9> in <module>() 1 val = 'a' 2 ----> 3 if val>0 and val<10: 4 print('Value: ', val, 'is a digit.') 5 elif val==0: TypeError: '>' not supported between instances of 'str' and 'int'
We can avoid problems like this by wrapping our code in an if statement:
if type(val) is int or type(val) is float:
if val>0 and val<10:
print('Value: ', val, 'is a digit.')
elif val==0:
print('Value ', val, 'is nul')
else:
print('Value: ', val, 'is a number.')
else:
print('val is not a number')
However, this becomes tedious to implement very quickly, especially when trying to cover a number of possible errors, which will all need their own if clauses. This can lead to a lot of duplicated code, making the code difficult to maintain.
Python provides the try-except structure to avoid this issue, enabling developers to properly manage any possible errors. The basic try-except structure is:
try:
if val>0 and val<10:
print('Value: ', val, 'is a digit.')
elif val==0:
print('Value ', val, 'is nul')
else:
print('Value: ', val, 'is a number.')
except:
print('Val is not a number')
print('Enter a new number')
At the top of the statement is the code that we are interested in executing, which is run in the try statement. If that fails then the except statement comes into effect, (hopefully) returning helpful information to the user about what happened and giving them some guidance on how to avoid the problem in future.
The except statement will catch all errors and so we do not, initially at least, need to know exactly what errors we are trying to avoid. However, python does provide error codes, which we can use to expand the structure to capture specific error types. For the example above, we would want to capture a TypeError:
try:
if val>0 and val<10:
print('Value: ', val, 'is a digit.')
elif val==0:
print('Value ', val, 'is nul')
else:
print('Value: ', val, 'is a number.')
except TypeError as err:
print('Val is not a number')
print('But our code does not crash anymore')
print('The run-time error is:', err)
As with if statements, multiple except statements can be used, each with a different error test. These can be followed by an (optional) catch-all bare except statement (as we started with) to catch any unexpected errors. Note that only one try statement is allowed in the structure.
try-except structures may also contain else and finally statements (following in order after the except statements). The else statement will be executed if no except statements are executed, while the finally statement will be executed after all other statements are finished.
try:
if val>0 and val<10:
print('Value: ', val, 'is a digit.')
elif val==0:
print('Value ', val, 'is nul')
else:
print('Value: ', val, 'is a number.')
except TypeError as err:
print('Val is not a number')
print('But our code does not crash anymore')
print('The run-time error is:', err)
else:
print('1/val = ', 1/val)
finally:
print('release memory')
del(val)
The typical use of the finally statement is to deal with the release of external resources (such as files or network connections) whether or not the attempted action has been successful.
More details about errors and exceptions can be found in the Python tutorials.
HTTP errors
Earlier we were introduced to the
requestslibrary. This is used to access web content, and returns a code indicating the request status. Theraise_for_statusfunction is provided for testing the request status, which returns arequest.HTTPErrorif there is an error. For example, if we request a missing webpage we get this error:import requests source_url='https://api.datacite.org/dois/10.48546/workflowhub.workflow.56.1000' response = requests.get(source_url) response.raise_for_status()Traceback (most recent call last): File "<ipython-input-108-06c23faa067a>", line 3, in <module> response.raise_for_status() File "/Users/mbessdl2/anaconda3/envs/myenv/lib/python3.9/site-packages/requests/models.py", line 943, in raise_for_status raise HTTPError(http_error_msg, response=self) HTTPError: 404 Client Error: Not Found for url: https://api.datacite.org/dois/10.48546/workflowhub.workflow.56.1000Write a
try-exceptstructure that usesraise_for_statusand deals with the error without crashing.Solution
import requests try: source_url='https://api.datacite.org/dois/10.48546/workflowhub.workflow.56.1000' response = requests.get(source_url) response.raise_for_status() except requests.HTTPError as err: print(err)404 Client Error: Not Found for url: https://api.datacite.org/dois/10.48546/workflowhub.workflow.56.1000
Assert
The try-except structure is useful for controlling small chunks of code, where we might reasonably expect that the user can fix the problem (where it is a program which takes a short time to run), or where our program itself will fix the problem (such as substituting NaN values for a failed calculation).
Sometimes, however, it is better to check the data before it is passed into the code and to trigger an early crash in the program, which returns a meaningful error. For example, this can be extremely useful in the case of large numerical calculations that take a long time to run and which will only use some of the provided variables after a significant amount of the computational work has already been carried out. In such situations your user will appreciate being warned early on that they have provided invalid data.
To conduct such a test we can use an assert statement. This follows the structure: assert <test>, <error message>. As an example of this, we can write the test for non-numerical values as:
val = 'a'
assert type(val) is float or type(val) is int, "Variable has to be a numerical object"
if val>0 and val<10:
print('Value: ', val, 'is a digit.')
elif val==0:
print('Value ', val, 'is nul')
else:
print('Value: ', val, 'is a number.')
Traceback (most recent call last):
File "<ipython-input-38-52d27526daab>", line 1, in <module>
assert type(val) is float or type(val) is int, "Variable has to be a numerical object"
AssertionError: Variable has to be a numerical object
Like the earlier if statement, this catches the problematic variable before it reaches our calculation. However it is far less intrusive code, and assert tests can be added in sequence, so that we can quickly and easily increase the coverage of such tests.
Testing for NaN’s (Not a Number)
Numpy provides Not a Number (nan) values, which can be used to indicate missing data. These are not caught by the test above:
import numpy as np val = np.nan assert type(val) is float or type(val) is int, "Variable has to be a numerical object" if val>0 and val<10: print('Value: ', val, 'is a digit.') elif val==0: print('Value ', val, 'is nul') else: print('Value: ', val, 'is a number.')Value: nan is a number.However numpy does provide the
isnan()function for testing if a value is NaN. Add an assertion test using this which will raise and error if the value is NaN.Solution
Because the test checks if a value is NaN, we need to add
notbefore the test, to ensure that theassertstatement fails if it is a NaN:import numpy as np val = np.nan assert type(val) is float or type(val) is int, "Variable has to be a numerical object" assert not np.isnan(val), "Variable must not be a NaN" if val>0 and val<10: print('Value: ', val, 'is a digit.') elif val==0: print('Value ', val, 'is nul') else: print('Value: ', val, 'is a number.')Traceback (most recent call last): File "<ipython-input-52-553a9c93f77a>", line 4, in <module> assert not np.isnan(val), "Variable must not be a NaN" AssertionError: Variable must not be a NaN
Key Points
try-exceptstructures are useful for catching errors as they occur
assertstructures are useful for forcing errors early, to avoid wasted effort
Units and Quantities
Overview
Teaching: 0 min
Exercises: 0 minQuestions
How can I ensure consistent units in my code?
Objectives
Creating quantity objects which include values and units
Applying unit conversions
Learn about different types of equivalence for units
Using units for annotating plots
Test different unit libraries
One of the most common problems in physics and the development of physical models is related to the conversion and management of physical units. Without clear management of physical units, and ensuring that the information passed between different parts of the system is consistent, accidents such as the Mars Climate Orbiter probe crash can occur.
In this lesson we will look at one of the available libraries in python for helping to avoid such problems. This is the units module in the astropy library.
Installing a new python library
Following the conda lesson, we install the astropy library using:
conda install astropy
The units module can then be imported in your scripts using:
from astropy import units as u
Importing as u is not necessary, but it is a common shorthand for this module, and so we’ll use it below.
Basics
Units can be added to a python variable by multiplying that variable by the unit:
length = 26.2 * u.meter
(note the use of US spellings for units).
When printing the variable the unit information will now be attached:
print(length)
The type of this new variable is an astropy Quantity:
type(length)
astropy.units.quantity.Quantity
You can still access the original object using value:
print(length.value)
type(length.value)
26.2
float
Quantities can be converted to other units, systems or factors, by using the function to(). For example:
length.to(u.km)
length.to(u.lightyear)
This enables the use of arithmetic operations when quantities have compatible units:
distance_start = 10 * u.mm
distance_end = 23 * u.km
length = distance_end - distance_start
print(length)
And it also enables the combining of quantities, for example, to calculate a speed:
time = 15 * u.minute
speed = length / time
print(speed)
By default the units library will select units to report for these values based on what the units are of the objects that you have passed it. You can, as before, convert these to the units of your choice:
print(speed.to(u.km/u.s))
You can also convert the units to the base (irreducible) units for the unit system you are using with the decompose function (changing the unit system choice will be covered later):
print(speed.decompose())
Choosing different base units
If you wish to reduce to different base units you can pass a list of those units to the
basesparameter when calling thedecomposefunction:print(speed.decompose(bases=['km','s']))Note that the order of the values in the list passed to
basesparameter doesn’t matter. However, the base units you choose must be either one of the original units used, or a standard base unit. This function cannot be used to convert from km to cm, for example. Instead it is useful where you only want to reduce select units within the object.
You can change the base system using functions such as si (to change to the default SI units) and cgs (to change to centimetre-grams-seconds units):
print(speed.cgs)
Reducing more complex quantities
We have been given some (slightly messy) code that calculates the jerk, or rate of change of acceleration, of a vehicle in the units
m/h/min^2:acceleration = (25 * u.meter) / (15 * u.minute) / (15 * u.minute) time_change = 1 * u.hour jerk = acceleration / time_change print(jerk)0.1111111111111111 m / (h min2)For the report we are writing we need to convert this to the units
km/hour^3, which of these changes will do this task for us?
jerk.decompose(bases=['km','hr'])jerk.to(u.km/u.hour**3)jerk.to(u.km/u.hour**2)Solution
Answer 2 is correct. Decompose will only reduce to base units (or existing units), and so can’t change minutes to hours, or metres to kilometres. Answer 3 won’t work, because we have to apply the modification to all units (including those which are already correct).
Imperial Units
The units library is focused on scientific notation. To use units from the (US) imperial system we can import the imperial library:
from astropy.units import imperial
These can then be used in the same manner as the standard units:
speed.to(imperial.mile/u.hour)
Equivalent Units
There are some unit conversions that would initially appear to be unconvertible. For example, it is possible to convert meters into Hertz. At first glance it seems to be wrong but if you know the quantities for wavelength and frequencies, it is indeed a valid conversion:
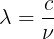 where:
where:
- lambda ~ m
- c ~ m/s
- ~ Hz ~ 1/s
This, however, does not work as above:
(656.281 * u.nm).to(u.Hz)
Instead we need to inform units what unit equivalency we want it to use, which can be specified with the equivalencies option:
(656.281 * u.nm).to(u.Hz, equivalencies=u.spectral())
In this case we use the spectral equivalence library, which allows conversions between wavelengths, wave number, frequency, and energy equivalent pairs. There are a number of other built-in equivalency libraries, for commonly used pairings.
Other built-in equivalencies are:
- Doppler (
dopplr_radio,doppler_optical,doppler_relativistic) - spectral flux density
- temperature
- brightness temperature
- temperature energy
- and you can build your own
Finding equivalencies is possible using the find_equivalent_units function:
u.Hz.find_equivalent_units()
Primary name | Unit definition | Aliases
[
Bq | 1 / s | becquerel ,
Ci | 3.7e+10 / s | curie ,
Hz | 1 / s | Hertz, hertz ,
]
This is a quite limited list - to see what is available for other equivalence libraries, we must specify these when making the request:
u.Hz.find_equivalent_units(equivalencies=u.spectral())
Primary name | Unit definition | Aliases
[
AU | 1.49598e+11 m | au, astronomical_unit ,
Angstrom | 1e-10 m | AA, angstrom ,
Bq | 1 / s | becquerel ,
Ci | 3.7e+10 / s | curie ,
Hz | 1 / s | Hertz, hertz ,
J | kg m2 / s2 | Joule, joule ,
Ry | 2.17987e-18 kg m2 / s2 | rydberg ,
cm | 0.01 m | centimeter ,
eV | 1.60218e-19 kg m2 / s2 | electronvolt ,
earthRad | 6.3781e+06 m | R_earth, Rearth ,
erg | 1e-07 kg m2 / s2 | ,
jupiterRad | 7.1492e+07 m | R_jup, Rjup, R_jupiter, Rjupiter ,
k | 100 / m | Kayser, kayser ,
lyr | 9.46073e+15 m | lightyear ,
m | irreducible | meter ,
micron | 1e-06 m | ,
pc | 3.08568e+16 m | parsec ,
solRad | 6.957e+08 m | R_sun, Rsun ,
]
Angles
The numpy trigonometry functions (np.sin, np.cos, etc) require angles that are passed to them to be in radians, rather than degrees. It is inconvenient to have to explicitly convert from other units to radians every time that you wish to use these functions, but with units you can have this done implicitly for you.
The radian unit is u.rad, and it’s equivalencies are:
u.rad.find_equivalent_units()
Primary name | Unit definition | Aliases
[
arcmin | 0.000290888 rad | arcminute ,
arcsec | 4.84814e-06 rad | arcsecond ,
cycle | 6.28319 rad | cy ,
deg | 0.0174533 rad | degree ,
hourangle | 0.261799 rad | ,
mas | 4.84814e-09 rad | ,
rad | irreducible | radian ,
uas | 4.84814e-12 rad | ,
]
We can see from this that the degree unit is u.deg, so we can use this to define our angles:
angle = 90 * u.deg()
print('angle in degrees: {}; and in radians: {}'.format(angle.value,angle.to(u.rad).value))
angle in degrees: 90.0; and in radians: 1.5707963267948966
Now we can pass the angle directly to np.sin without having to convert directly to radians:
print('sin of 90 degrees is: {}'.format(np.sin(angle)))
print('sin of pi/2 radians is: {}'.format(np.sin(1.57079632)))
print('sin of 90 degrees is not: {}'.format(np.sin(90)))
sin of 90 degrees is: 1.0
sin of pi/2 radians is: 1.0
sin of 90 degrees is not: 0.8939966636005579
Plotting Quantities
Support has been added for using the units information in matplotlib plots. To use this we need to import the quantity_support library:
from astropy.visualization import quantity_support
We will create a numpy array of the angles between 0-180 degrees, and plot the sin of these:
angles = np.arange(181) * u.deg
plt.plot(angles, np.sin(angles))
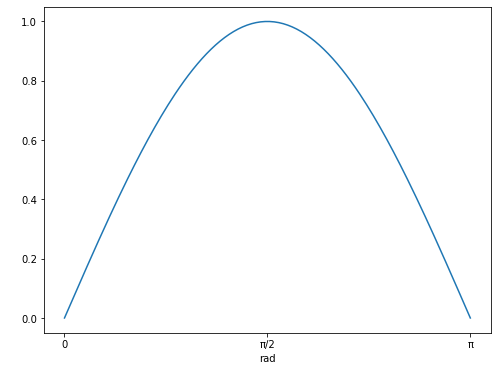
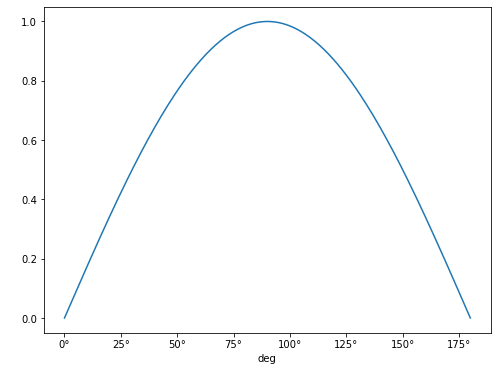 Note that the units for the x-axis are properly presented. This can be done for any angular unit we wish:
Note that the units for the x-axis are properly presented. This can be done for any angular unit we wish:
plt.plot(angles.to(u.rad), np.sin(angles))
Temperature
Each of the temperature scales is considered as using an irreducible unit in standard usage. As a consequence of this we need to always specify that we are using the u.temperature equivalences when we convert between these:
t2 = 1 * u.deg_C
print('{} is equivalent to {}'.format(t2,t2.to(imperial.Fahrenheit, equivalencies=u.temperature())))
1.0 deg_C is equivalent to 33.8 deg_F
But, be aware that these are scales, if we wanted to increase a temperature given in Fahrenheit by 1 degree Celsius, we would not add 33.8 degrees Fahrenheit!
To get the incremental value of 1 degree Celsius in Fahrenheit we would need to do this:
t1 = 0 * u.deg_C
t2 = 1 * u.deg_C
print('{} increment is equivalent to a {} increment'.format(
t2 - t1,
t2.to(imperial.Fahrenheit,equivalencies=u.temperature())
- t1.to(imperial.Fahrenheit, equivalencies=u.temperature())
)
)
1.0 deg_C increment is equivalent to a 1.7999999999999972 deg_F increment
This is verging on unreadable (as is demonstrated by us having to split a simple arithmetic expression involving two variables across two lines of code).
Fortunately there is support for cleaner temperature conversions in the pint package. This has similar functionality to the astropy.unit package, but has been built from the ground up with a focus on different aspects. These include unit parsing and standalone unit definitions, uncertainties integration, and (important for our example here), cleaner treatment of temperature units.
Pint can be installed using conda (and the conda-forge channel):
conda install -c conda-forge pint
The unit registry is loaded using:
from pint import UnitRegistry
ureg = UnitRegistry()
A units definition file could be given here if needed, but in this case we will stick with the default settings.
This library has the temperature units as before:
t2 = 1 * ureg.degC
print('{} is equivalent to {}'.format(t2,t2.to(ureg.degF)))
1 degree_Celsius is equivalent to 33.79999999999993 degree_Fahrenheit
But it also includes the concept of temperature increments:
deltaT = 1 * ureg.delta_degC
print('{} is equivalent to {}'.format(deltaT,deltaT.to(ureg.delta_degF)))
1 delta_degree_Celsius is equivalent to 1.7999999999999998 delta_degree_Fahrenheit
Because of the popularity of python this overlap of functionality of packages can be common. We would recommend exploring the different available packages that might cover your requirements before settling on one. And keep watching out for new packages, and be prepared to switch when starting new projects if/when you find better packages.
Key Points
astropy.unitslibrary provides unit support
Quantityobjects are created by multiplying values by the desired unitsThe
.to()function can be used to convert unitsThe
.decompose()function can be used to convert to the base (irreducible) unitsEquivalences can be found using the
.find_equivalent_units()functionSpecific equivalence libraries can be defined using the
equivalences=keywordImport quantity-support from
astropy.visualizationto integrate units with matplotlib for data plottingThe
pintlibrary provides similar unit support, but is better for working with temperature increments
Pandas Essential
Overview
Teaching: 0 min
Exercises: 0 minQuestions
How can I work with spreadsheet style datasets?
Objectives
Learn to load csv data, and obtain simple analyses of the data
Learn how to index data arrays using column/row labels
Learn how to mask data to remove unwanted values
Learn how to plot data directly from pandas
This library is based on the numpy library that we discuss in a previous lesson. It provides python users with a new object that allows us to work with “relational” or “labelled” data in an easy way. It provides similar access to data loaded from databases or spreadsheets as those found in languages like R.
Pandas is designed more for data science than pure numerical analysis but the required tools can easily be combined together. In addition, the library provides input/output tools that allow the opening and saving of data from MS Excel, CSV or HDF5 files.
The library is usually imported as pd:
import pandas as pd
Later we will need to plot data - so the matplotlib library should be loaded:
import matplotlib.pyplot as plt
We will be working with data in the data/gapminder_gdp_europe.csv file. Open this
with a text editor and look at the data layout.
The data within this file is organised much as you’d expect the data within a spreadsheet. The first row of the file contains the headers for each of the columns. The first column contains the name of the countries, while the remaining columns contain the GDP values for these countries for each year. Pandas has the read_csv function for reading structured data such as this, which makes reading the file easy:
data = pd.read_csv('data/gapminder_gdp_europe.csv',index_col='country')
Here we specify that the country column should be used as the index column (index_col).
This creates a DataFrame object containing the dataset. This is similar to a numpy array, but has a number of significant differences. The first is that there are more ways to quickly understand a pandas dataframe. For example, the info function gives an overview of the data types and layout of the DataFrame:
data.info()
<class 'pandas.core.frame.DataFrame'>
Index: 30 entries, Albania to United Kingdom
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 gdpPercap_1952 30 non-null float64
1 gdpPercap_1957 30 non-null float64
2 gdpPercap_1962 30 non-null float64
3 gdpPercap_1967 30 non-null float64
4 gdpPercap_1972 30 non-null float64
5 gdpPercap_1977 30 non-null float64
6 gdpPercap_1982 30 non-null float64
7 gdpPercap_1987 30 non-null float64
8 gdpPercap_1992 30 non-null float64
9 gdpPercap_1997 30 non-null float64
10 gdpPercap_2002 30 non-null float64
11 gdpPercap_2007 30 non-null float64
dtypes: float64(12)
memory usage: 3.0+ KB
You can also carry out quick analysis of the data using the describe function:
data.describe()
gdpPercap_1952 gdpPercap_1957 gdpPercap_1962 ...
count 30.000000 30.000000 30.000000 ...
mean 5661.057435 6963.012816 8365.486814 ...
std 3114.060493 3677.950146 4199.193906 ...
min 973.533195 1353.989176 1709.683679 ...
25% 3241.132406 4394.874315 5373.536612 ...
50% 5142.469716 6066.721495 7515.733738 ...
75% 7236.794919 9597.220820 10931.085347 ...
max 14734.232750 17909.489730 20431.092700 ...
Accessing elements, rows, and columns
The other major difference to numpy arrays is that we cannot directly access the array elements using numerical indices such as data[0,0]. It is possible to access columns of data using the column headers as indices (for example, data['gdpPercap_1952']), but this is not recommended. Instead you should use the iloc and loc methods.
The iloc method enables us to access the DataFrame as we would a numpy array:
print(data.iloc[0,0])
while the loc method enables the same access using the index and column headers:
print(data.loc["Albania", "gdpPercap_1952"])
For both of these methods, we can leave out the column indexes, and these will all be returned for the specified index row:
print(data.loc["Albania"])
This will not work for column headings (in the inverse of the data['gdpPercap_1952'] method) however. While it is quick to type, we recommend trying to avoid using this method of slicing the DataFrame, in favour of the methods described below.
For both of these methods we can use the : character to select all elements in a row or column. For example, to get all information for Albania:
print(data.loc["Albania", :])
or:
print(data.iloc[0, :])
The : character by itself is shorthand to indicate all elements across that indice, but it can also be combined with index values or column headers to specify a slice of the DataArray:
print(data.loc["Albania", 'gdpPercap_1962':'gdpPercap_1972'])
If either end of the slice definition is omitted, then the slice will run to the end of that indice (just as it does for : by itself):
print(data.loc["Albania", 'gdpPercap_1962':])
Slices can also be defined using a list of indexes or column headings:
year_list = ['gdpPercap_1952','gdpPercap_1967','gdpPercap_1982','gdpPercap_1997']
country_list = ['Albania','Belgium']
print(data.loc[country_list, year_list])
gdpPercap_1952 gdpPercap_1967 gdpPercap_1982 gdpPercap_1997
country
Albania 1601.056136 2760.196931 3630.880722 3193.054604
Belgium 8343.105127 13149.041190 20979.845890 27561.196630
Masking data
Pandas data arrays are based on numpy arrays, and retain some of the numpy tools, such as masked arrays. This enables us to apply selection criteria to the datasets, so that only the values that we require are shown. For example, the following selects all data where the GDP is above $10,000:
subset = data.loc['Italy':'Poland', 'gdpPercap_1962':'gdpPercap_1972']
print(subset[subset>10000])
gdpPercap_1962 gdpPercap_1967 gdpPercap_1972
country
Italy NaN 10022.40131 12269.27378
Montenegro NaN NaN NaN
Netherlands 12790.84956 15363.25136 18794.74567
Norway 13450.40151 16361.87647 18965.05551
Poland NaN NaN NaN
Plotting from Pandas
Pandas is integrated with matplotlib, and so data can be plotted directly using the integrated plot method. For example, to plot the GDP for Sweden:
data.loc['Sweden',:].plot()
plt.xticks(rotation=90)
 Note that we’ve had to rotate the xtick labels by 90 degrees, because they do not fit neatly under the x-axis. Later we will clean these up properly.
Note that we’ve had to rotate the xtick labels by 90 degrees, because they do not fit neatly under the x-axis. Later we will clean these up properly.
Note that, in the case above, we passed a single column of data to the plot method - which it automatically transposed in order to make sense of the plot request (because this method usually works on rows of data). If you want to plot more than one column of data you will need to explicitly transpose the DataFrame yourself.
For example, we will transpose the GDP data for the first 3 countries in our dataset:
print(data.iloc[0:3,:].T)
country Albania Austria Belgium
gdpPercap_1952 1601.056136 6137.076492 8343.105127
gdpPercap_1957 1942.284244 8842.598030 9714.960623
gdpPercap_1962 2312.888958 10750.721110 10991.206760
gdpPercap_1967 2760.196931 12834.602400 13149.041190
gdpPercap_1972 3313.422188 16661.625600 16672.143560
gdpPercap_1977 3533.003910 19749.422300 19117.974480
gdpPercap_1982 3630.880722 21597.083620 20979.845890
gdpPercap_1987 3738.932735 23687.826070 22525.563080
gdpPercap_1992 2497.437901 27042.018680 25575.570690
gdpPercap_1997 3193.054604 29095.920660 27561.196630
gdpPercap_2002 4604.211737 32417.607690 30485.883750
gdpPercap_2007 5937.029526 36126.492700 33692.605080
This data is now ready to be plotted as a histogram - first we set the style of the plot to match that of the ggplot package in R:
plt.style.use('ggplot')
data.iloc[0:3,:].T.plot(kind='bar')
plt.xticks(rotation=90)
plt.ylabel('GDP per capita')
Saving figures created by pandas
Some python interpreters will, when creating a figure, present you with a graphical interface that you can use to save the figure. If, however, your interface does not do this (or you are writing a script which will automatically generate and save the figures), then you will first need to define a matplotlib figure object, and ensure that pandas plots your figure inside this.
To do this create a matplotlib subplots object, with handles for the figure (
fig) and axis (axs) objects. Pass the axis object to pandas when plotting your figure (ax=axs), then use the figure object at the end to save the figure (fig.savefig).fig, axs = plt.subplots() data.loc['Albania':'Belgium',:].T.plot(kind='bar',ax=axs) plt.xticks(rotation=90) plt.ylabel('GDP per capita') fig.savefig('albania-austria-belgium_GDP.png', bbox_inches='tight')
Changing Column Labels
Note that the x-tick labels have been taken directly from the index values of the transposed DataFrame (which were the original column labels). These don’t really need to be more than the year of the GDP values, so we could change the column labels to reflect this.
First we make a new copy of the dataframe (in case anything goes wrong):
gdpPercap = data.copy(deep=True)We have given this new dataframe a more appropriate name, replacing the information that will be removed from the column headers.
Now we will use the inbuilt
str.stripmethod to clean up our column labels for the new dataframe. Which of these commands is correct:
gdpPercap.columns = data.columns.str.strip('gdpPercap_')gdpPercap = data.columns.str.strip('gdpPercap_')Solution
The correct answer is 1. We have to pass the new column labels explicitly back to the array columns, otherwise all we do is replace the data array with a list of the new column labels.
Plotting GDP increases with time
Now that we’ve cleaned up the column labels, we now want to plot the GDP data for Sweden and Iceland from 1972 onwards. The code block we will be using is:
gdp_percap<BLOCK>.T.plot(kind='line') # Create legend. plt.legend(loc='upper left') plt.xlabel('Year') plt.ylabel('GDP per capita ($)')Which of the following blocks of code should replace the
<BLOCK>in the code above?
.loc['Sweden':'Iceland','gdpPercap_1972':].loc['gdpPercap_1972':,['Sweden','Iceland']].loc[['Sweden','Iceland'],'gdpPercap_1972':].loc['gdpPercap_1972':,'Sweden':'Iceland']Solution
The correct answer is 3. The two countries are not adjacent in the dataset, so we need to use a list to slice them, not a range (disqualifying answers 1 and 4). At the point where we select the countries using
.loc, we have not yet transposed the dataset (using.T), so the country names are still indexes, not column labels, and therefore need to be referenced first (ie in the first set of square brackets), (disqualifying answers 2 and 4).
Key Points
CSV data is loaded using the
load_csv()functionThe
describe()function gives a quick analysis of the data
loc[<index>,<column>]indexes the data array by the index and column labels
iloc[<index>,<column>]indexes the data array using numerical indiciesThe data can be sliced by providing index and/or column indicies as ranges or lists of values
The built-in
plot()function can be used to plot the data using thematplotliblibrary